Laboratory for Informatics, Networks and Systems (LINS)

Data mining and machine learning:
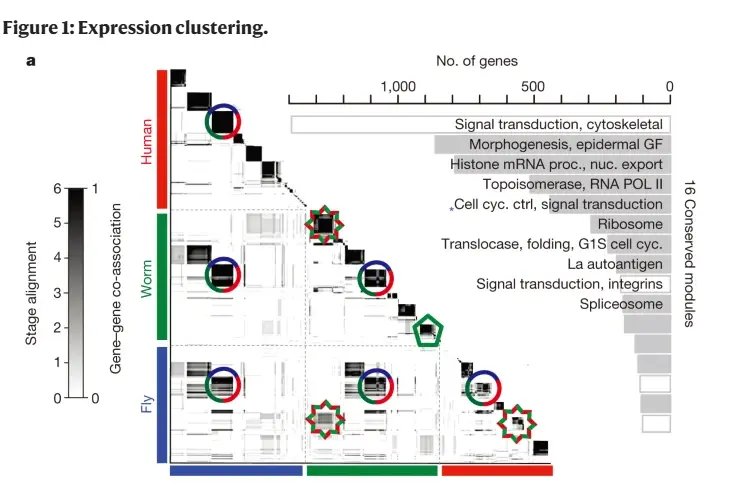
Data mining and machine learning: we designed a novel cross-species clustering algorithm to demonstrate conserved and species-specific gene and non-coding RNA regulatory modules during embryonic development between C. elegans and D. mel. We found that in both species, the orthologous genes work more closely during the phylotypic developmental stage (aka the vertebrate body plan stage) than other developmental stages. This lays the groundwork for evolutionary expression patterns during embryogenesis and enabled us to systematically study interactions between evolutionary conserved and species-specific functions during development [Nature 512, 445–448, 2014; Genome Biology 15:R100, 2014].
Computational and mathematical modeling:
Computational and mathematical modeling: we developed computational methods identifying the principal gene expression patterns for complex biological processes such as embryogenesis, allowing integration of the state-space model and dimensionality reduction by matrix factorizations for the first time. This approach produced an entirely new analytical platform with promise to open new avenues of investigation into systematic and robust dynamic patterns from high dimensional, complex and noisy gene expression data [PLoS Computational Biology, 12(10): e1005146, 2016; PLoS ONE 7(1): e28805, 2012; IEEE/ACM Transactions on Computational Biology and Bioinformatics, 430-437, 2012].
Systems biology:
Systems biology: we developed a computational method by integrating ENCODE and TCGA data to identify a genome-wide regulatory logic of transcription factors and microRNAs reporting on logic patterns observed in leukemia. Until this point, similar logics had only been reported in simple organisms like yeast. These results provided unprecedented insights into the gene regulatory circuit logics in complex and more advanced biological systems like cancer [PLoS Computational Biology 11(4): e1004132, 2015].

Translational science:
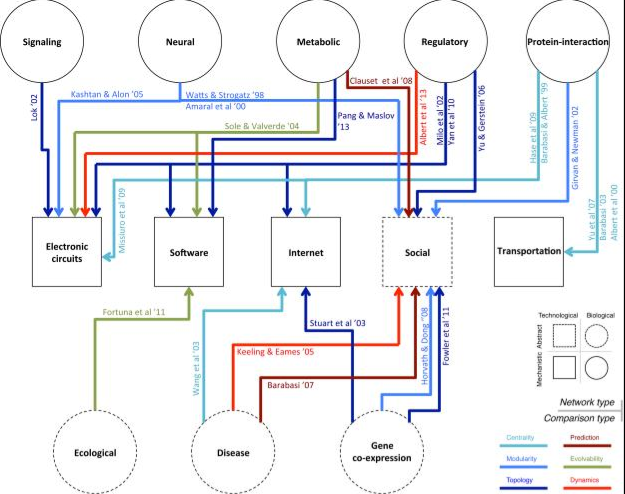
Translational science: our recent review compared the characteristics of biological networks with other disciplines, and discussed the cross-disciplinary transferability of network formalisms to help gain novel biological insights at the system level. We illustrated how these comparisons benefit the field with a few specific examples related to network growth, organizational hierarchies, and the evolution of adaptive systems [Cell Systems, 2, 147-157, 2016].
Network science:
Network science: we analyzed the academic social networks driven by large scientific consortia (Big Science), which revealed temporal dynamics of collaborative patterns between consortia members and non-member users [Trends in Genetics, 32, 251-253, 2016].

What is Machine Learning in Genomics?
Machine Learning uses algorithms to identify patterns and make predictions based on data, without being explicitly programmed for specific tasks. In genomics, ML helps to analyze high-dimensional datasets generated by sequencing technologies.
What is Network Science?

Gene Co-expression Networks:
Identifying genes with correlated expression to infer functional relationships.
Protein-Protein Interaction (PPI) Networks:
Understanding cellular mechanisms by mapping interactions between proteins.
Pathway Analysis:
Studying metabolic and signaling pathways to identify critical genes or proteins.
Disease Networks:
Mapping genetic variants to disease phenotypes through interaction networks.
Network Science studies the structure and dynamics of networks (graphs) that model relationships between entities. In genomics, these entities can be genes, proteins, or regulatory elements, and their interactions form complex biological networks.
Integration of Machine Learning and Network Science in Genomics
The combination of ML and Network Science enables the analysis of complex, large-scale genomic datasets to uncover relationships and mechanisms that are otherwise challenging to discern.

Graph Neural Networks (GNNs):
- An ML method tailored for network data, where genes, proteins, or other biological entities form nodes, and their interactions form edges.
- Applications include predicting node properties (e.g., gene essentiality) or edge existence (e.g., protein interactions).

Community Detection & Clustering:
- Network Science methods to find modules or clusters in genomic networks.
- ML can enhance clustering by incorporating additional features like gene expression data or sequence motifs.

Feature Extraction from Networks:
Networks provide topological features (e.g., centrality measures) that can be used as inputs for ML models to predict gene or protein roles.

Dynamic Network Modeling:
Studying temporal changes in genomic networks (e.g., during disease progression) using ML to model dynamics.

Network-Based Feature Selection:
Using network topology to prioritize features for downstream ML tasks like classification or regression.